Peptide Library & Pool Design
JPT has been designing and synthesizing peptide libraries and peptide pools for more than 15 years. Each design facilitates answering specific scientifc questions.
We are happy to provide recommendations to choose the most efficient peptide library or pool design for your specific application.
Common Peptide Library & Pool Designs
Positional Scanning Library
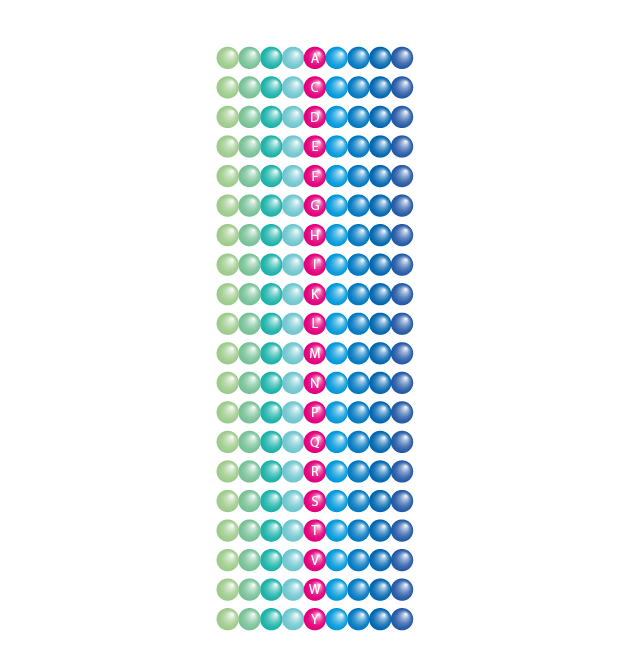
- Increase activity of enzyme substrates
- Enhance antibody epitopes
- Improve T-cell epitopes
- Optimize peptide binding sites
Overlapping Peptide Library
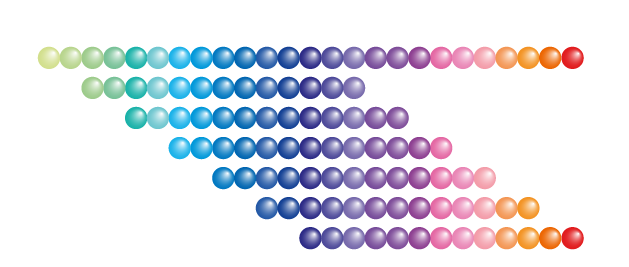
- Scan an antigen sequence for epitopes (epitope mapping)
- Screen a protein for a substrate
- Search for other binding sites within a given protein
- Identify a T-cell epitope
- Stimulate T-cells in T-cell assays
- Map antibody epitopes
Neo-Epitope Library
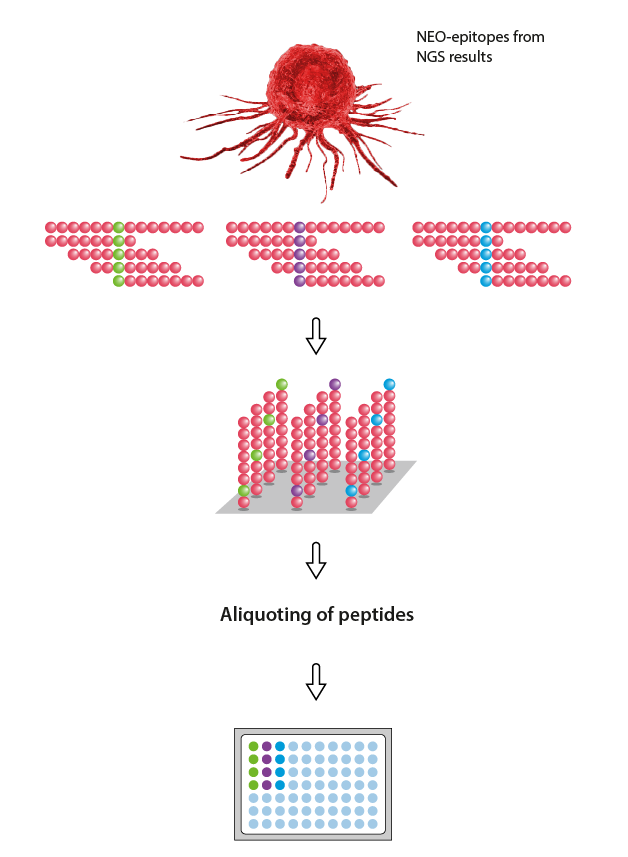
Use our free and easy-to-use PepSequencer for generation of overlapping peptide sequences from neo-epitopes or contact us!
Related Products
Peptide Modifications & Specialty PeptidesSpotMix Antigen Discovery Pools
NeoSpikeMix for Immunopeptidomics
Cyclization Library
- Development of peptides with enhanced bio stability
- Mimicking secondary protein structures (e.g. protein loops)
- Optimizing peptides (e.g. peptide ligands with increased binding potency/selectivity and enhanced protease stability)
Related Products
Antigen Discovery Pool
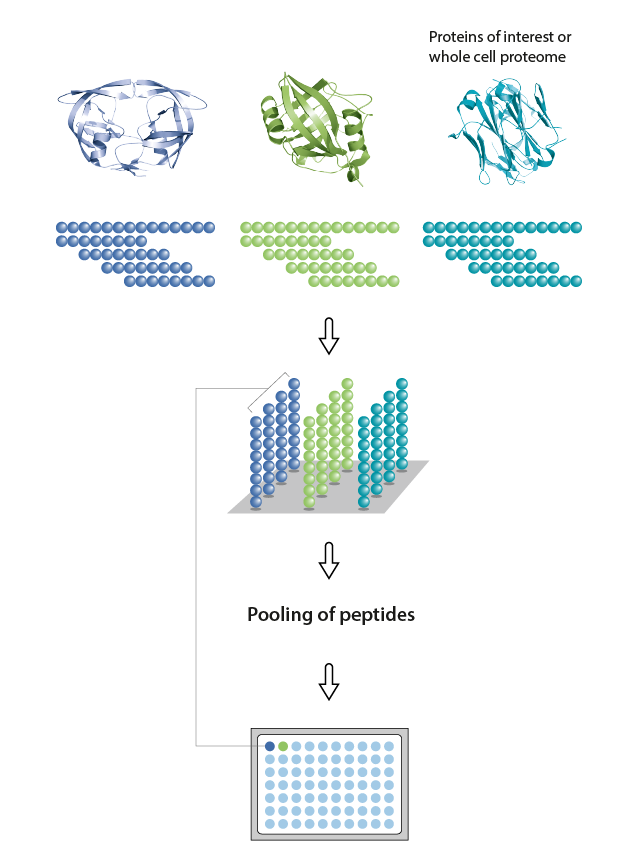
- Epitope discovery
- Screening projects
- Target discovery
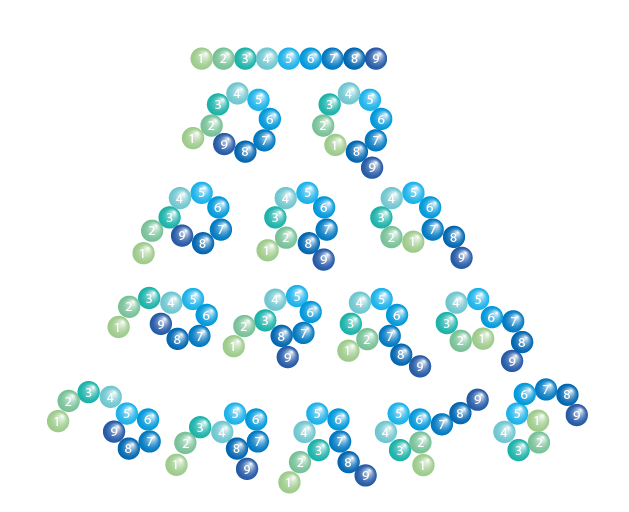
Matrix Pools
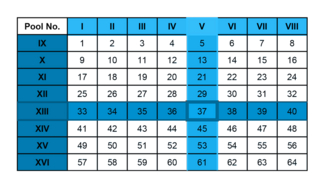
Matrix Pools have a specialized layout that enables fast and minimal material consuming identification of epitopes within an antigen. From a given antigen overlapping peptides are generated and pooled into many subpools.
Each peptide is present in exactly two subpools according to a layout that we design for each antigen.
In the example shown below 64 peptides are pooled into 16 Matrix Pools. Pools V and XIII elicit a positive T-cell response. Only peptide 37 is present in both pools and therefore is the peptide containing the epitope.
Matrix Pools are also the main component of Epitope Mapping Peptide Sets (EMPS).
Contact us for further information!
ULTRA Library & Pools
ULTRA libraries and pools are used to represent sequence diversity with the least possible number of peptides for
- Immune monitoring of T- and B-cell response
- T-cell stimulation in immunotherapy
- Biomarker discovery
- Development of vaccines
We generate ULTRA libraries with our proprietary algorithm to reflect the full spectrum of sequence variation within a virus or type of cancer with one comprehensive peptide library. This saves precious time and resources.
Our algorithm creates all possible peptide variations and scores these according to their frequency of occurrence across all sequences. We can then define the optimal overlap required to provide homogeneous overall coverage.
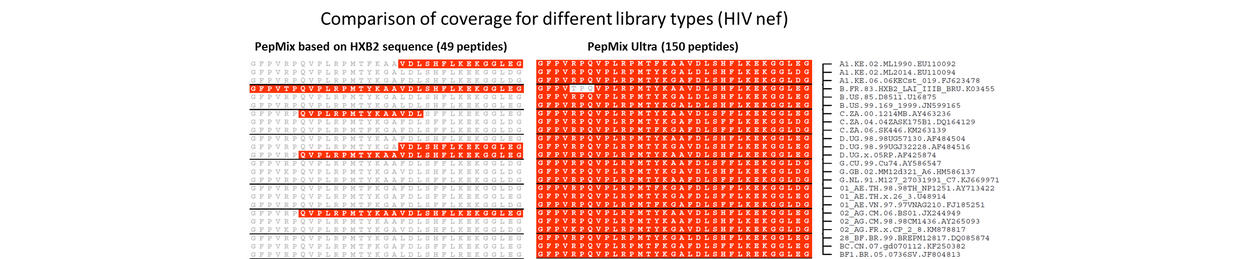
Sequence Diversity in Viruses
- Hallmark in many pathogenic viruses including HIV, HBV, Influenza and SARS-CoV-2
- Most sequence diversity found in viral envelope proteins, due to their important role in eliciting host immune recognition
- Sequence diversity has to be taken into account in the design of antigen-based products, intended for immune monitoring and vaccination
Sequence Diversity in Tumors
- 10 000 to 12 000 sites with coding mutations have been identified per individual, translating into a sequence alteration in one of every two genes
- Somatic mutations cause sequence heterogeneity within individuals which are of special significance in the pathological mechanisms of cancer
- 767 germline and 29881 somatic mutations have been identified for TP53, one of the most important tumor driver genes
Selected Epitope Pool
Selected Epitope Pools consist of a number of peptides representing key immunodominant epitopes from antigens or pathogens. Peptides are selected from pre-identified epitopes with respect to HLA restrictions.
Well known examples are our CEFX Ultra SuperStim and CEF control pools that contain epitopes from different organisms and the Pan Select Pools containing all immunodominant epitopes from one virus each.
The benefits of selected epitope pools are
- Well characterized epitopes provide a representative picture of the T cell response
- Smart selection of epitopes creates powerful controls for T-cell standardization
- Individual peptide responses can be mapped more easily
- Efficient and cost effective method for classifying T cell response
Request a price for your customized peptide pool.
Truncation Library

A truncation library is used to
- Determine the minimum length of an epitope (minimal epitope)
- Map the minimal binding site of a protein that is required for proper binding
Truncation libraries are generated by reducing the peptide length (truncation) amino acid per amino acid from each terminus creating a set of truncated peptides with different lengths. If the key residues have already been identified using an alanine scanning library , truncation can be stopped at the key residues position.
Related Products
Alanine Scanning Library
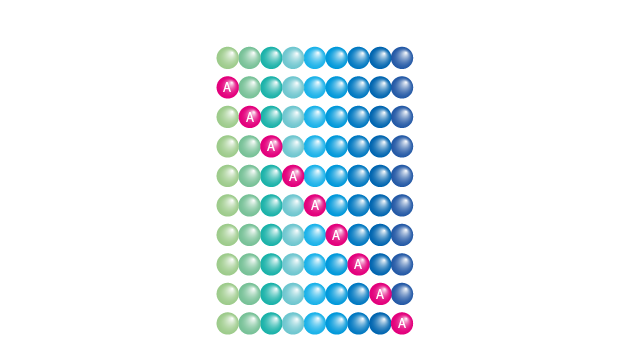
Alanine scanning libraries are used to
- Identify key residues in epitopes
- Search binding parameters of enzymes
- Study binding sites of proteins
Creating an alanine scanning library, alanine systematically substitutes the residues at each amino acid position of the original peptide i.e. identified epitope (= alanine walk). Substitution of an important or essential amino acid by alanine results in less or no binding or activity.
Related Products
Random Library

A random library is used for peptide sequence optimization. To generate random peptide libraries selected residues of a peptide sequences (wobbles) are randomly and simultaneously substituted with all or some of the 20 natural amino acids via a shotgun approach. Alternatively, a random library with specific physico-chemical properties can be designed by computer scientists resulting in random peptide libraries of defined size and properties for target identification and drug discovery.
Contact us for the generation of random libraries!
Related Products:
Scrambled Library
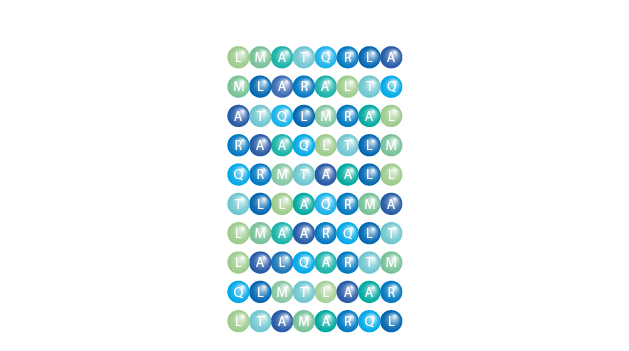
Scrambled Libraries are used for screening proteins of interest, e.g. antibodies or enzymes with the permutations of a peptide for sequence optimization. A scrambled library is generated by permutation of the original peptide sequence and represents all alternative peptides with the same amino acid composition. Scrambled peptides or scrambled peptide libraries are also used as negative controls.
Contact us for the generation of scrambled libraries!
Related Products
Looking for further peptide knowledge?
