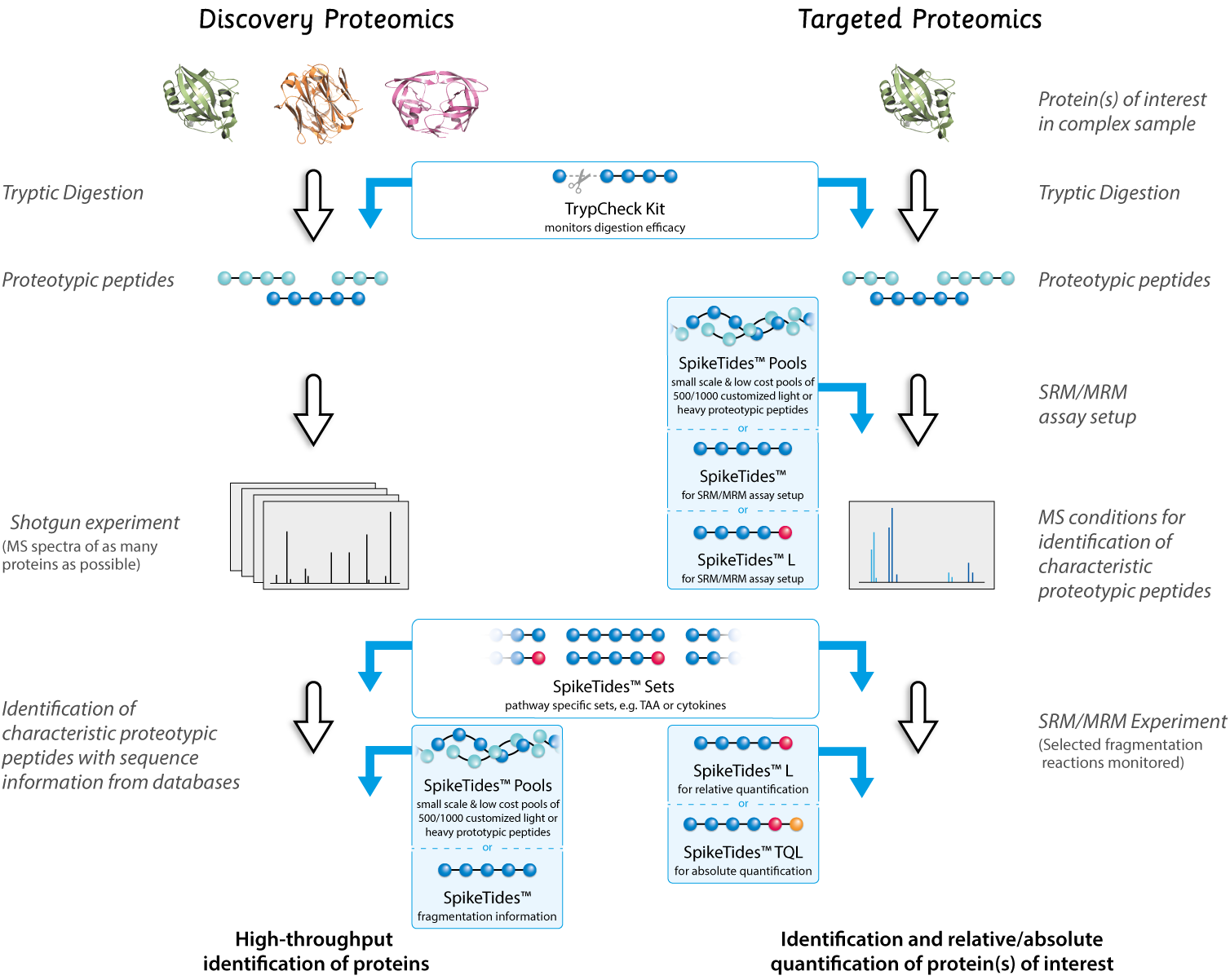
Mass Spectrometry Based Proteomics
Proteomics is the study of a cell's protein inventory at different times by protein identification and quantification. The application of LC-MS/MS has tremendously facilitated this process. Generally, samples containing relevant proteins are digested into peptides and analyzed by LC-MS/MS as a surrogate measurement of protein levels. To increase accuracy of detection and to enable protein quantitation, chemically synthesized stable isotope-labeled (SIL) peptides are added to samples. As traditional assembly and quantitation methods for such peptides are tedious and expensive, JPT has developed unique and highly efficient procedures for both fast and low-cost synthesis as well as absolute quantitation of SIL peptides.
SpikeTides™ & SpikeMix™ Reference Peptides for Targeted Proteomics
- Inexpensive & fast delivery (10 000 peptides/ week)
- For targeted proteomics (protein detection and protein quantification by mass spectrometry)
- Monitoring of cellular regulation by incorporation of post-translational modifications
Proteomics Standards
- Normalize HPLC-MS retention time
- Compare collision energy settings
- Monitor efficacy of carbamidomethylation
- Check efficacy of trypsination
- Standardize MRM assays across species
Absolute Quantified Peptides SpikeTides™ TQL
Protein Interaction Screen on Peptide Matrix (PRISMA)
- Identification of protein-protein interaction partners
- Decoding the influence of post-translational modifications
- Comparison of cell states
- Mapping of pathways
NeoSpikeMix for Immunoproteomics
- Hundreds or thousands of peptides within one week
- Low price
- Provides high sensitivity as needed for low abundance epitopes
PTM Peptide Reference Standards