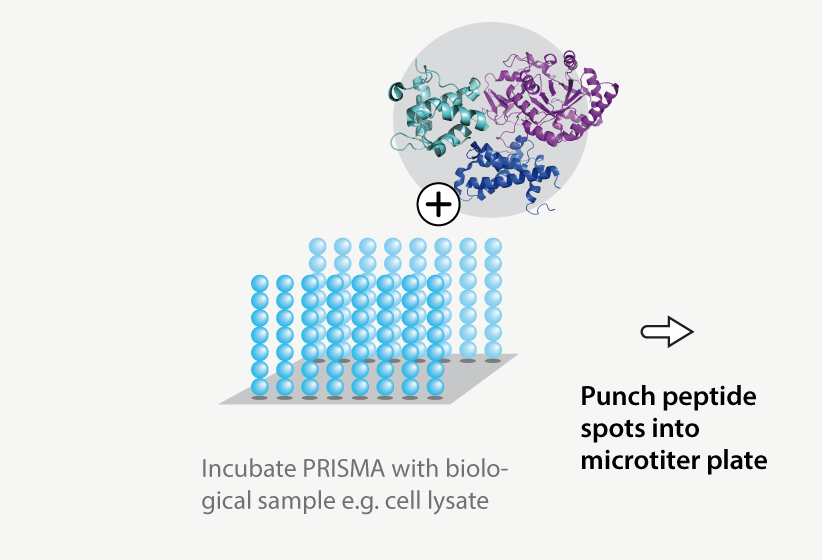
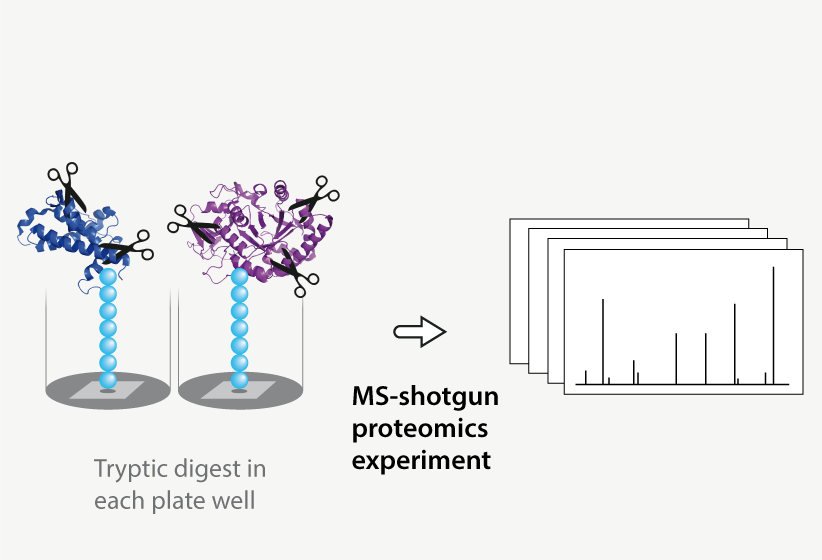
Protein-protein interactions are a hallmark of signal transmission and key to understanding regulatory mechanisms. Therefore, we developed the Protein Interaction Screen on Peptide Matrix (PRISMA) together with our partners from Max-Delbruck-Center for Molecular Medicine (MDC) to systematically explore the interactome.
- Peptides are synthesized on a cellulose membrane
- Peptides of up to 25 amino acids can represent sequence diversity and contain post-translational modifications
- The membrane is incubated with a biological sample for affinity enrichment of soluble proteins and protein complexes
- Proteins interacting with the peptide matrix are then identified and quantified by mass spectrometry


Quality Assurance
All production is ISO 9001:2015 certified.