Partner with JPT
JPT Peptide Technologies is an ISO 9001:2015 certified innovative service provider and research & development partner. Our R&D services and cooperations leverage our know how in peptide technologies to advance projects in various applications such as
- Vaccine target and epitope discovery
- Immune monitoring and diagnostic content research
- Seromarker discovery and validation
- Systematic peptide optimization
- Discovery, clinical and functional proteomics
Our Capabilities
Why partner with JPT?
- Proprietary technologies for peptide synthesis, analysis and high throughput screening
- In-house R&D team for organic and medicinal chemistry, immunology and assay development, structural biology, bioinformatics and data management
- Solid track record of successful R&D projects
- Flexible business arrangements
- ISO 9001:2015 regulated quality management system
- Professional project organisation and communication
What do we offer?
- Fee-for-service projects
- FTE based and milestone driven collaboration projects
- Shared risk, partnered development projects
- Participation on publicly granted R&D projects
Track record
- Peptide selection and synthesis for individualized neo-epitope based vaccination strategies
- Peptide pool design and synthesis for adoptive cell transfer in gene therapy
- Design of comprehensive peptide libraries for diagnostics and treatment
- Identification of serological predictive, prognostic, and surrogate biomarkers
- Proteome-spanning target discovery for vaccine development
- T-cell epitope discovery
- Development of next generation reference standards for targeted proteomics
- Design and synthesis of peptide libraries for proteome wide detection and quantification of proteins
- Development of molecular probes for the multiplexed detection and quantification of proteins from biological samples
- Design and synthesis of screening libraries
- Identification and optimization of agonistic and antagonistic ligands
- Identification and optimization of affinity ligands
- Substrate identification and optimization for orphan enzymes
Applications
Immunotherapy & Vaccines Drug Discovery & Optimization Proteomics
Projects
JPT Peptide Technologies is an ISO 9001:2015 certified research & development partner for peptide-based projects. Our proprietary peptide technologies help to advance development projects for a variety of applications such as novel immune monitoring tools and diagnostic content, seromarker discovery and validation, vaccine target discovery and peptide lead identification, systematic peptide optimization of peptide structures as well as proteomics standards.
Here we present a selected number of projects:
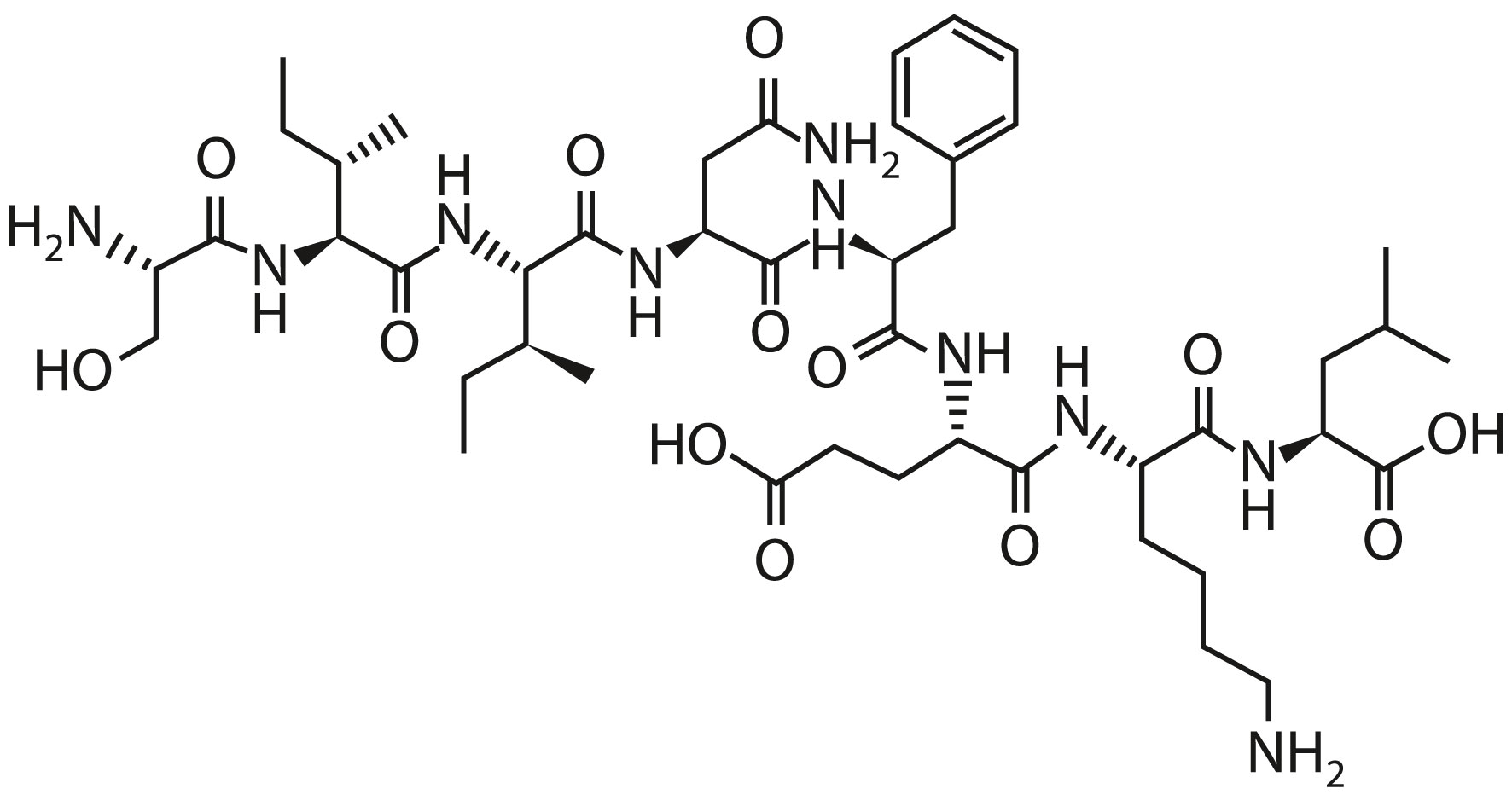
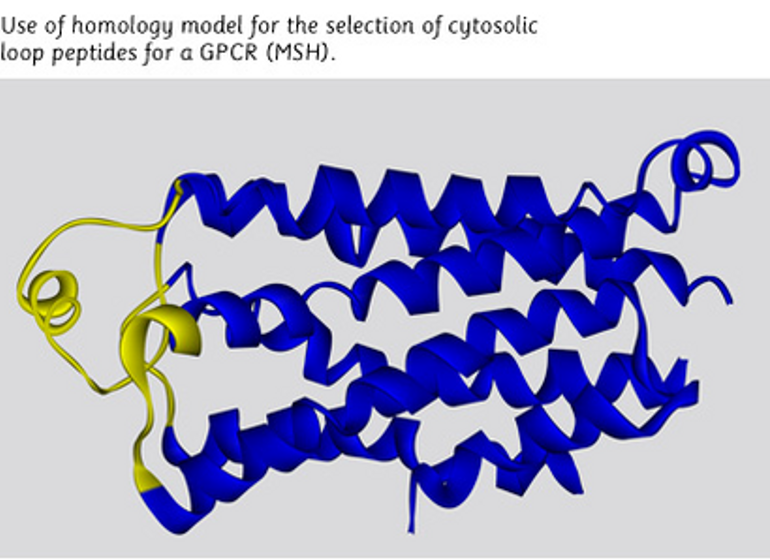
SARS-CoV-2 T Cell Immunity SARS-CoV-2 B Cell Immunity Peptide Mimotope Optimization